#!/usr/bin/env python
# coding: utf-8
# # Machine Learning with Python
#
# Collaboratory workshop
#
# This is a notebook developed for the second day of the Collaboratory Workshop, Machine Learning with Python. For more information, go to the workshop home page:
#
# https://github.com/kose-y/W17.MachineLearning/wiki/Day-2
# - __Day 1__ - Fundamentals and Motivation
# - __Day 2__ - Classification and Cross-Validation
# - Assessing the performance of your models
# - Learning to choose and use models from Scikit-learn
# - Cross validation
# - __Day 3__ - Regression and Unsupervised Learning
# Open Jupyter Notebook and load `numpy` and `matplotlib.pyplot`.
# ## Installing an additional package (Optional, for local installations)
#
# Let's install another package for today's workshop: `graphviz`. This package is for visualizing graphs (or networks), a data structure consisting of nodes and edges connecting the nodes. In this workshop, it will be used for visualizing decision trees.
#
# If you are on Windows, open `Anaconda Prompt`. Ohterwise, open a terminal (also accesible on JupyterLab). Then type:
#
# ```
# conda install python-graphviz
# ```
#
# This is a simple way to install an anaconda-supported package.
#
# _Note_: This is already done for Binder environment.
# Now, let's load packages for today's workshop:
# In[ ]:
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams["font.size"] = 15 # increase font size in the figure
# ## Loading the synthetic data
# Let's consider a simple but useful example:
#
#

#
#

#
#

#
#
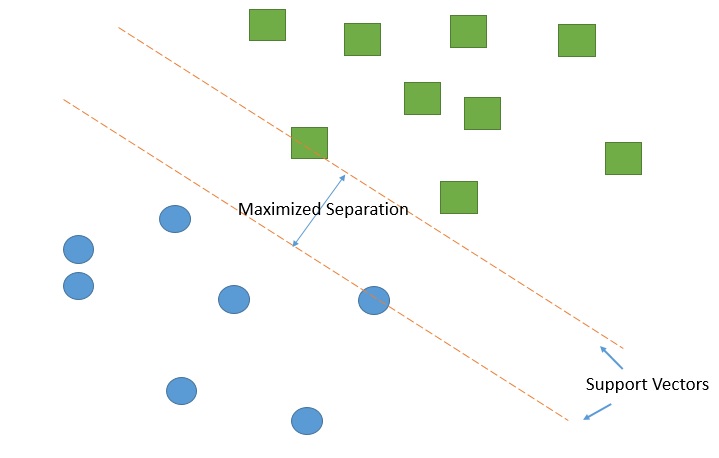
#
#

#
#

#
#

#
 #
#  #
#  #
#  #
#  #
#  #
#  #
#  #
#